Eleonora De Lazzari* published her findings on scaling laws for gene families. A striking quantitative invariant in evolutionary genomics is the scaling with genome size of the number of proteins sharing a specific function. E.D.L. showed that such scaling laws exist systematically at the level of single evolutionary families. This provides a novel view of the links between evolutionary expansion of protein families and gene functions.This work was performed in collaboration with J Grilli (U. Chicago) and S Maslov (U. Illinois).
|
|
|
|
|
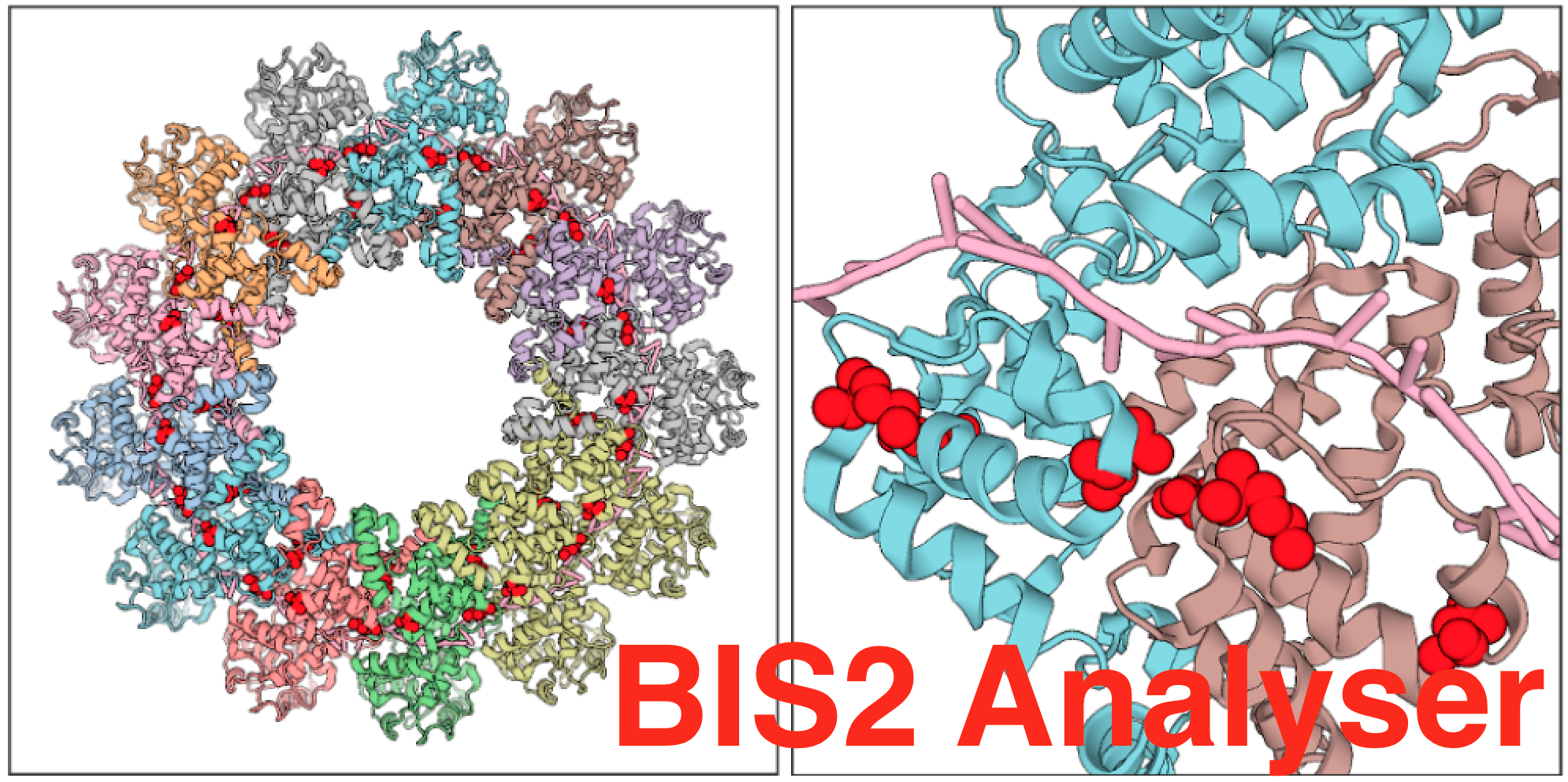
BIS2Analyzer is a webserver providing the online analysis of coevolving amino-acid pairs in protein alignments, especially designed for vertebrate and viral protein families, which typically display a small number of highly similar sequences. (A.Carbone team)
To the article
|
Plasmobase is a unique database designed for the comparative study of 11 Plasmodium genomes. Plasmobase proposes new domain architectures as well as new domain families that have never been reported before for these genomes. It allows for an easy comparison among architectures within Plasmodium species and with other species, described in UniProt. Joint work of J.Bernardes and A.Carbone.
Server
To the article
|
Angela Falciatore and Andrés Ritter organize the Algae workshop at 8th annual meeting EFOR network, May 2nd and 3rd 2017 Paris
|
Alessandra Carbone is interviewed by "Binaire - L'informatique: la science au coeur du numérique", the Le Monde Blog dedicated to the impact of computer science in our society (in collaboration with TheConversation). The interview was conducted by Serge Abiteboul and Claire Mathieu.
To the Article
|
Marco Cosentino-Lagomarsino and Gilles Fischer collaborated with Gianni Liti’s team to generatate end-to-end genome assemblies for 12 yeast genomes based on long-read sequencing. These population-level high-quality genomes with comprehensive annotation enable the first explicit definition of chromosomal boundaries between core and subtelomeric regions as well as a precise quantification of their relative evolutionary rates of genome dynamics.
To the article
|
|
|
The Diatom Functional Genomics Team (Angela Falciatore and Antonio E. Fortunato) contributed to the manuscript “Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus”, published on Nature in January 2017. This study, coordinated by Prof. Thomas Mock, University of East Anglia, Norwich, has revealed the existence of highly diverged alleles in the genome of this polar diatom species that may be involved in adaptation to environmental fluctuations in the Southern Ocean.
To the Article
|